BrAPI High Dimensional Phenotypes
Expanding the digital ecosystem for high-dimensional phenotypes in plant breeding
High dimensional phenotypes (HDPs) are promising tools for the advancement of diverse biological research, especially in plant breeding and genetics. HDPs, also known as “omics” data, encompass a variety of data types, such as spectral, metabolomic, transcriptomic, proteomic, and microbiome metagenomic data. These data types can inform our collective knowledge of biological processes and enable more efficient plant breeding selection decisions.
HDPs share a characteristic structure in which each observation is comprised of an annotated vector of sub-observations (e.g., individual metabolite abundances for metabolomics). This complex structure poses challenges for HDP storage, transfer, and analysis.
This project aims to increase the accessibility of HDPs for plant breeding programs by developing HDP BrAPI endpoints and BrAPI-enabled collection, management, and analysis tools that facilitate the integration of new HDP data types with traditional plant breeding data.
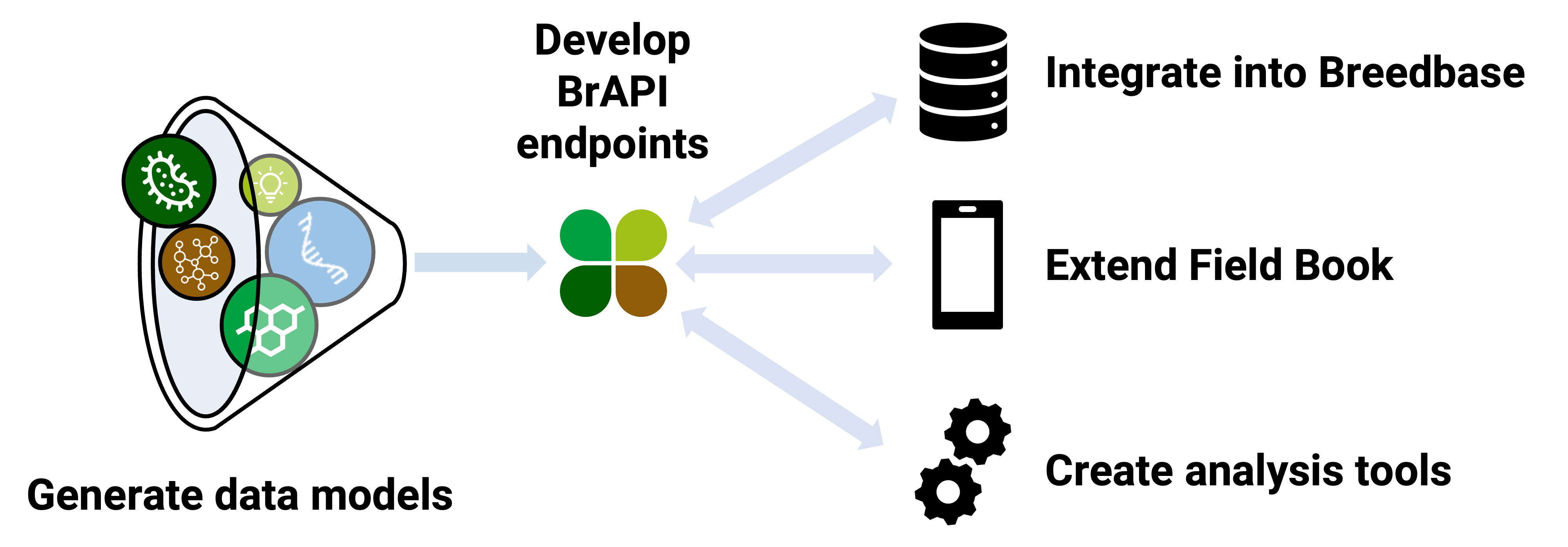
Approach
- Generate appropriate data models for HDPs (spectral, transcriptomic, metabolomic, proteomic, and microbiome data) that accurately represent data structure and associated metadata for plant breeding and genetics use cases.
- Develop BrAPI standards to efficiently handle HDPs based on the data models for each data type.
- Design and implement HDP storage structures in BrAPI-enabled breeding databases.
- Integrate HDP BrAPI calls into widely-used plant breeding data collection tools.
- Develop HDP BrAPI-enabled analysis applications (BrAPPs) that integrate omics and other plant breeding data types.
Team
- Jenna Hershberger, Clemson University - PD
- Trevor Rife, Clemson University - co-PI
- Lukas Mueller, Boyce Thompson Institute - co-PI
- Jean-Luc Jannink, USDA-ARS - collaborator
- Peter Selby, Cornell Univeristy - collaborator
Funding
This work is supported by the Agriculture and Food Research Initiative Data Science for Food and Agricultural Systems, project award no. SC-2023-11724 from the U.S. Department of Agriculture’s National Institute of Food and Agriculture.